This article introduces the preprocessing of EEG and EMG data. The goal is to analyze the correlation between EEG and EMG data on the same timescale, and the raw data collected is in TXT format with EEG and EMG data stored together. You can directly use the graphical interface here: Github Repo: EEG_EMG Process
This article covers the processing of single data. If you use my graphical UI interface, you can process data in batches. It will automatically read data from files for processing.
Before preprocessing the EEG data, understanding the basic format of the data is crucial. Below is an example of the format of the collected EEG and EMG data:
1 | # MIX|None|0+True+胫骨前肌+1000|1+True+腓骨长肌+1000|2+True+腓肠肌内侧+1000|3+True+腓肠肌外侧+1000|4+True+股直肌+1000|5+True+股内侧肌+1000|6+True+股二头肌长头+1000|7+True+半腱肌+1000|8+False+胫骨前肌+1000|9+False+腓骨长肌+1000|10+False+肠肌内侧+1000|11+False+肠肌外侧+1000|12+False+EMG13+1000|13+False+EMG14+1000|14+False+EMG15+1000|15+False+EMG16+1000|0+True+P4+80|1+True+CP2+80|2+True+FC5+80|3+True+C3+80|4+True+P3+80|5+True+C2+80|6+True+FC6+80|7+True+C4+80|8+True+CP6+80|9+True+F3+80|10+True+FC2+80|11+True+FC1+80|12+True+F4+80|13+True+CP5+80|14+True+C1+80|15+True+CP1+80 |
MIX|None|: Describes the data type (e.g., MIX could be mixed signals).X+True/False+Channel+SampleRate: Represents information about different channels, including whether they are enabled (True/False), the channel name (e.g., Tibialis Anterior, P4, etc.), and the sampling rate (in Hz).#, such as 13425+24732+39459.... This represents marker data from the experiment. However, in this experiment, markers are not used due to inaccuracies.If you use my graphical interface for processing, you don’t need to worry about this step—it will be done automatically.
First, use MATLAB to read the raw data from the file with the following code:
1 | fid = fopen(filepath); |
Based on the experimental requirements, extract the EEG and EMG data. EMG is from columns 1-8, and EEG is from columns 17-32.
1 | EMGData = [datafile{1:8}]; |
1 | function [EMGData, EEGData] = filterData(EMGData, EEGData) |
Due to missing or inaccurate marker information, you need to manually correct the marker data. First, plot the data and find the precise positions from the graph.
1 | N1 = length(EMGData(1,:)); % Number of EMG channels |
You can manually recover the original positions after obtaining the dotting information, or store them separately.
You need to manually construct the event information file.
Event information files must use plain text format (.txt), where each line represents a single event record, and fields are separated by spaces, commas, or tabs. It is recommended to use tab-separated (tab-separated values) for easy parsing.
| Field Name | Data Type | Required | Description |
|---|---|---|---|
type |
String | Yes | Event type identifier, e.g., 'Stimulus' or 'Response'. |
latency |
Numeric | Yes | The sampling point of the event in the EEG data (in samples). |
duration |
Numeric | No | Duration of the event in samples, fill with 0 if unknown. |
epoch |
Numeric | No | The segment number, required only when EEG data is segmented. |
urevent |
Numeric | No | The event number linked to the original event data for traceability. |
The file can be stored simply as events.txt, for example:
1 | type latency duration |
At this step, cut the data based on the dotting information and the duration of each event, leaving only the useful parts.
set)1 | EEG = pop_importdata('dataformat','ascii','nbchan',0,'data',filepath,'setname',eventFileName,'srate',1000,'pnts',0,'xmin',0,'chanlocs',locs_path); |
Parameter Explanation:
| Parameter | Example Value | Description |
|---|---|---|
dataformat |
'ascii' |
Data format type; 'ascii' means the data is stored in text format, other values can be 'matlab' or native formats. |
nbchan |
0 |
Number of channels in the data, 0 means auto-detect the number of channels (common), or you can specify the actual count. |
data |
filepath |
Path to the data file, which contains actual signal data; each row represents a time point, and columns represent channel data. |
setname |
eventFileName |
Name of the dataset that will appear in the EEGLAB graphical interface, used to identify the imported EEG dataset. |
srate |
1000 |
Sampling rate of the data in Hz. For example, 1000 means 1000 samples per second. |
pnts |
0 |
Number of samples per channel, 0 means auto-detected from the file. |
xmin |
0 |
The start time of the data (in seconds); 0 means from the first sample point. |
chanlocs |
locs_path |
The path to the channel location information file, typically .loc or .ced files, which define the spatial locations (electrode positions). |
My chanlocs file looks like this:
1 | Number labels theta radius X Y Z sph_theta sph_phi sph_radius type |
Note: The original file did not look like this. However, when directly importing raw EEG, the position may rotate 90 degrees, leading to an error. It requires manual correction each time the data is imported, or once the positions are fixed, export the corrected map and store it for future use.
pop_eegplot to draw EEG waveform graphs.1 | pop_eegplot(EEG, 1, 1, 0); % Draw the waveform for the data |
EEG: The EEGLAB data structure, containing the imported data.1 (continuous signal mode): Plot the raw continuous signal.1 (channel index label): Display channel labels on the plot.0 (single window): Display all channels in a single window; 1 would show each channel in a separate window.Plotting via the graphical interface
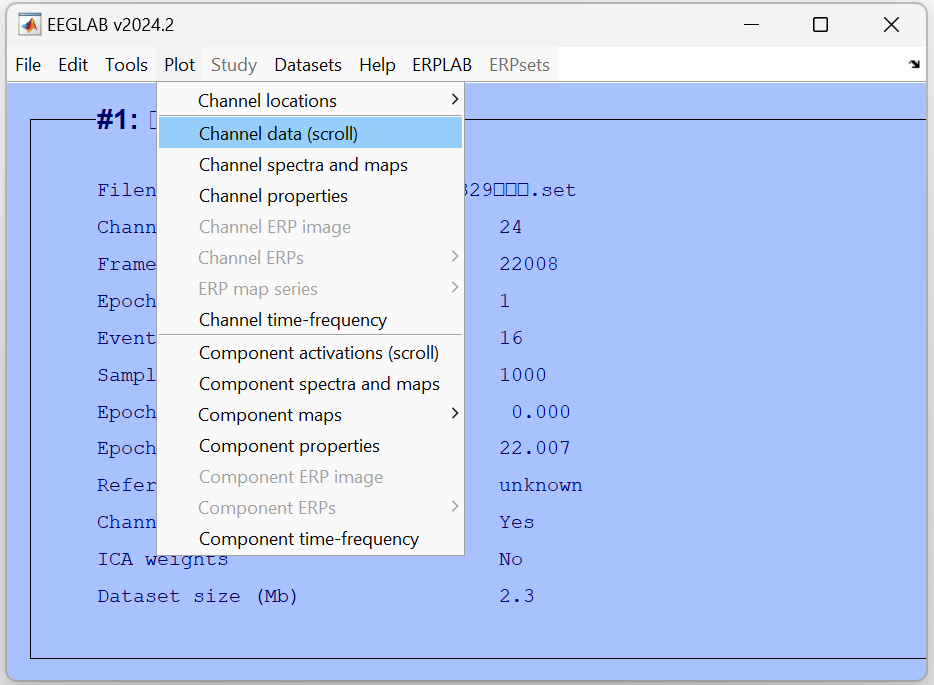
Based on the waveform plot, determine which channels are corrupted by checking:
Signal Anomalies:
Spatial Inconsistency:
Check the head map for the electrode distribution. A damaged channel may appear isolated on the map with abnormal signal spikes/drops in contrast to neighboring channels.
1 | EEG_interp = pop_interp(EEG, selectedIndices, 'spherical') |
Parameter Explanation:
EEG: The original EEG data structure.selectedIndices: An array of indices for channels requiring interpolation.'spherical': The interpolation method. This spherical interpolation takes into account the three-dimensional spatial arrangement of electrodes on a sphere, which is appropriate for EEG data.Manual Interpolation
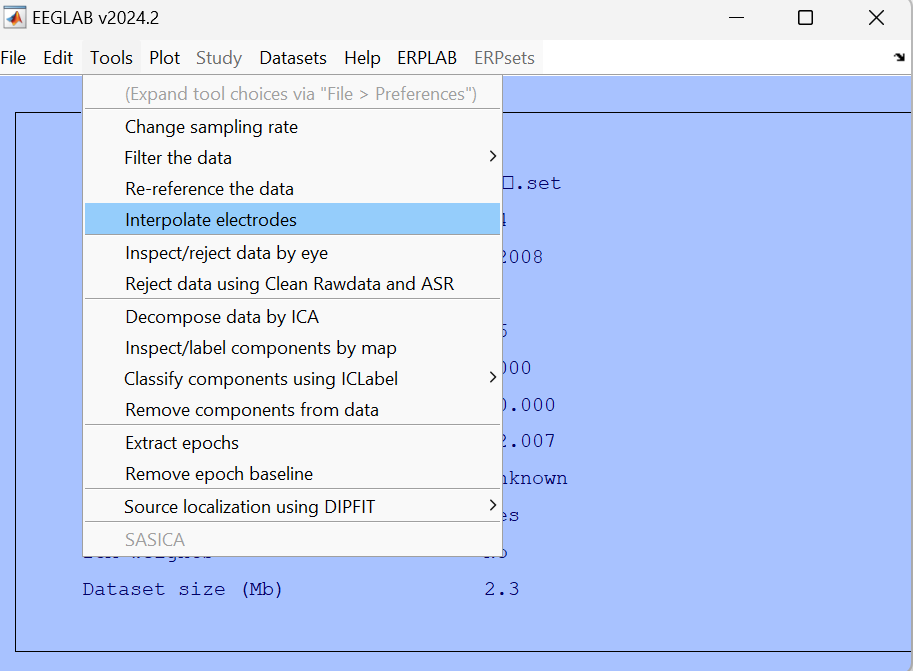
Select the channels that need interpolation.
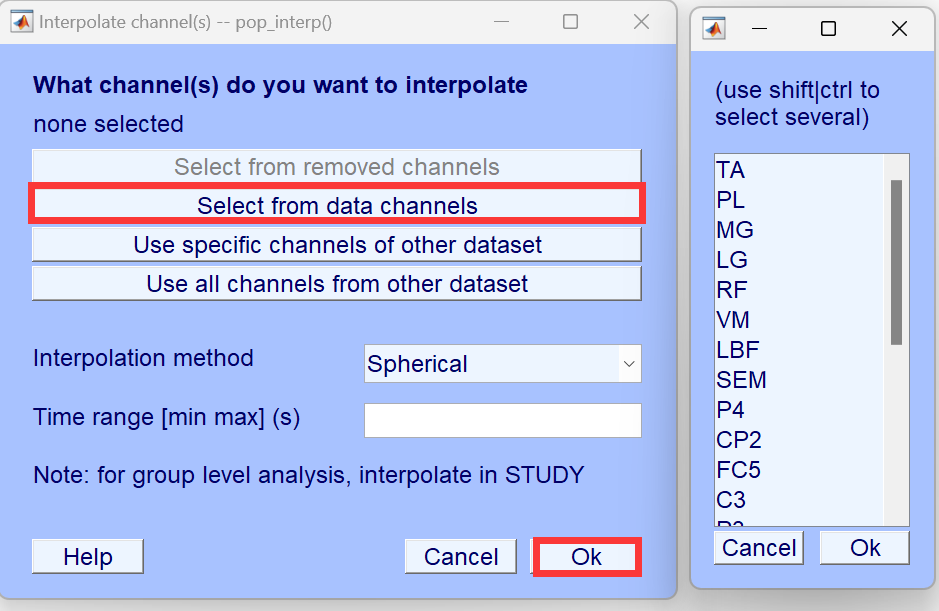
Note: Interpolation must be done for all channels at once, and cannot be performed in multiple steps.
To remove artifacts such as eye movements or muscle activities, use ICA:
1 | EEG = pop_runica(EEG, 'icatype', 'runica', 'extended', 1, 'interrupt', 'on', 'chanind', EEG_chans, 'pca', restLen); |
Parameter Explanation:
icatype, runica: The ICA algorithm to be used (RunICA).extended, 1: Use extended mode, which adds dimensionality to the analysis, suitable for multichannel data.interrupt, on: Allow interruption during ICA computation.chanind, EEG_chans: Indices of EEG channels that will participate in ICA (since EEG and EMG data are together, we should select the EEG channels).pca, restLen: Using PCA to reduce the data’s dimensions. restLen represents the number of principal components (EEG electrode count - interpolated bad channel count).This step aims to extract independent components from EEG data to eliminate artifacts while retaining components that reflect neural activity.
Or you can use the manual approach.
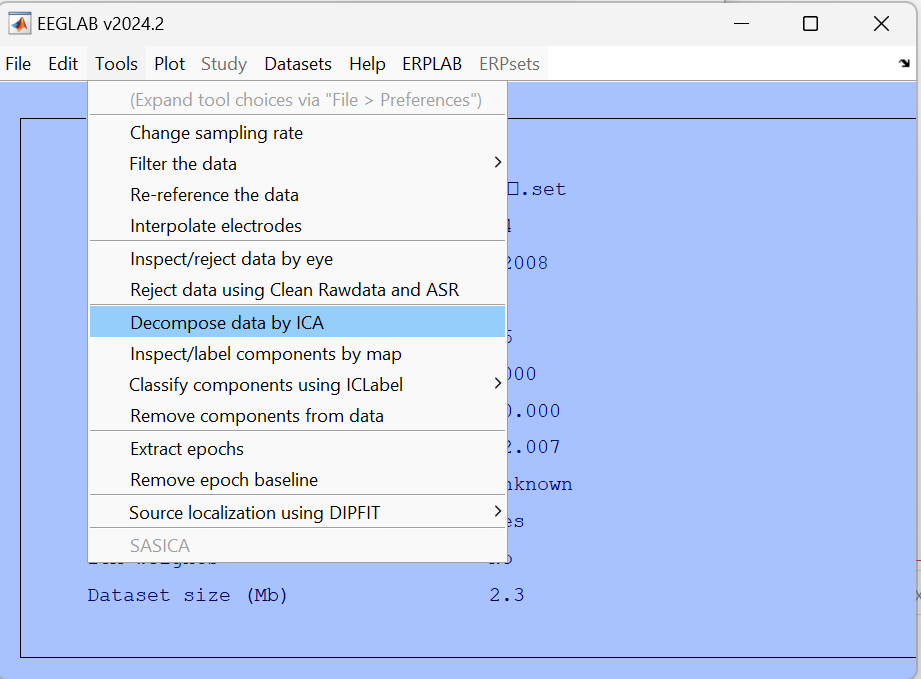
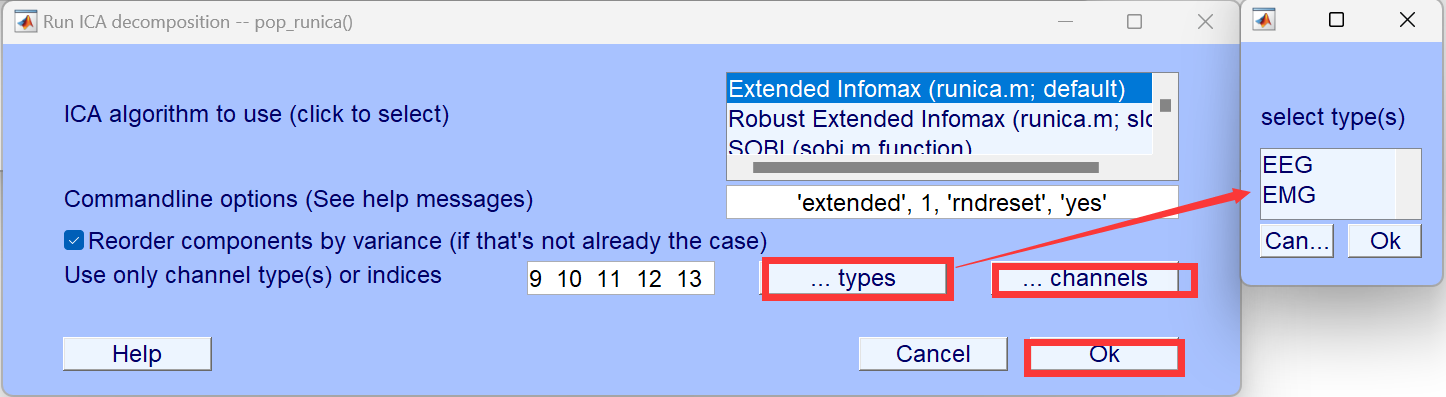
The data selection can be done in two ways, assuming categories for channels are already labeled.
During the execution, windows can pop up for real-time interruptions if necessary.
You can mark the data with labels, tagging artifacts as necessary:
1 | EEG_temp = EEG; % |
Manual execution: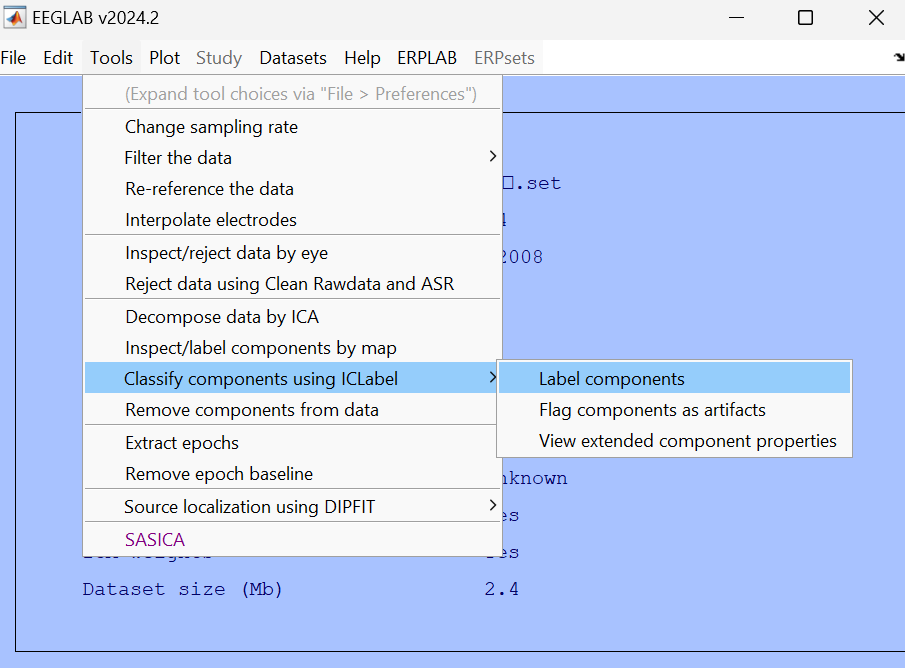
After marking, the flagged content will pop up. This image doesn’t serve any particular purpose and can be closed directly; it’s the classification drawn after automatic artifact analysis.
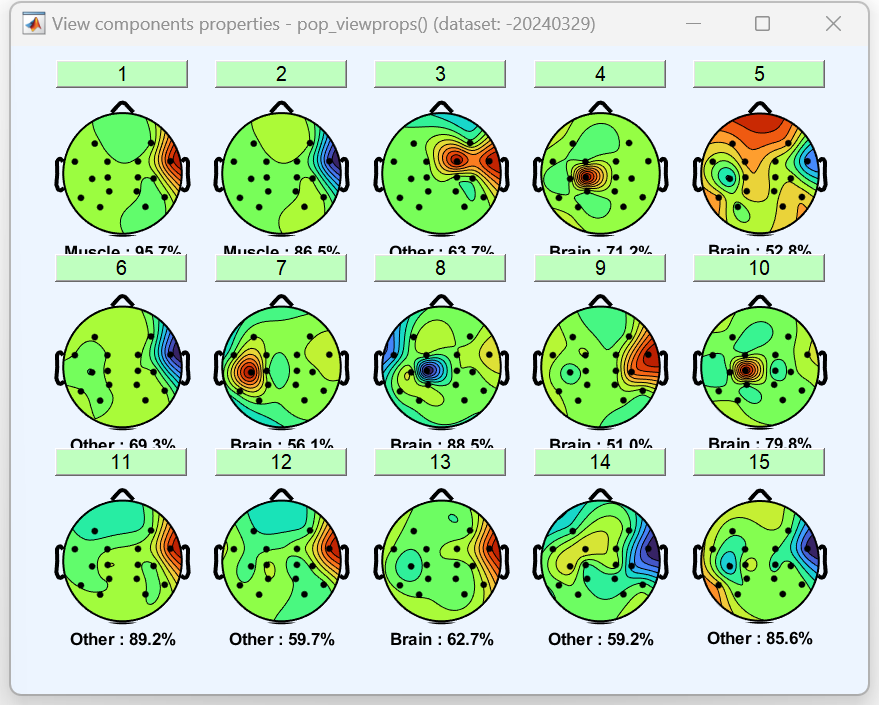 Then use the "flag components as artifact" function to automatically mark the artifacts.
Then use the "flag components as artifact" function to automatically mark the artifacts.
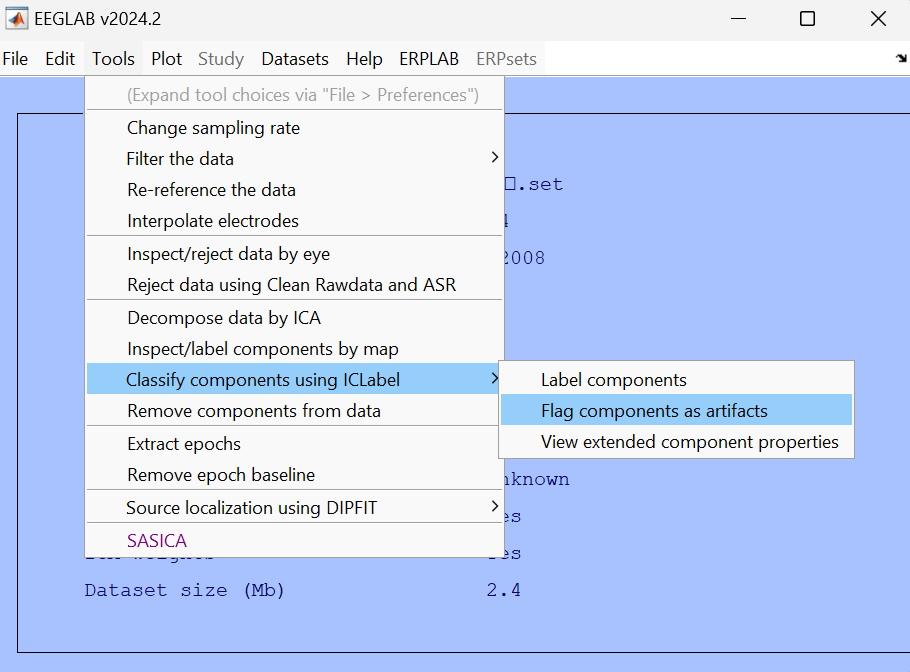 Set thresholds for different categories.
Set thresholds for different categories.
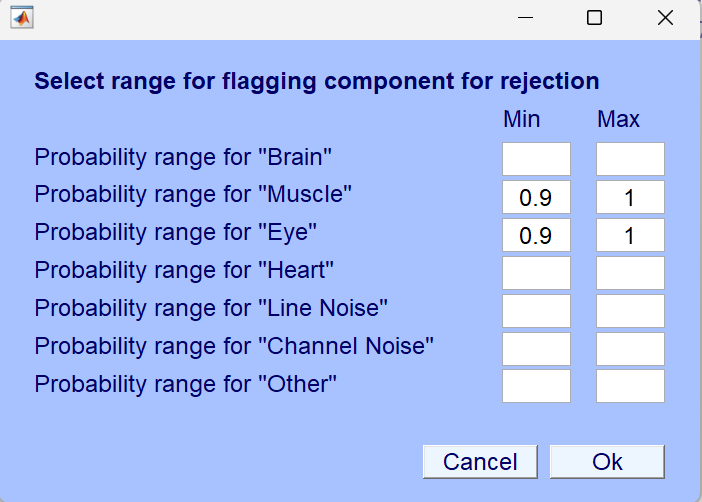 Set the required thresholds based on your data. For my own data, I set the threshold for each category to 0.55, because my data has many interference factors.
Set the required thresholds based on your data. For my own data, I set the threshold for each category to 0.55, because my data has many interference factors.
1 | rejected_comps = find(EEG_temp.reject.gcompreject > 0); |
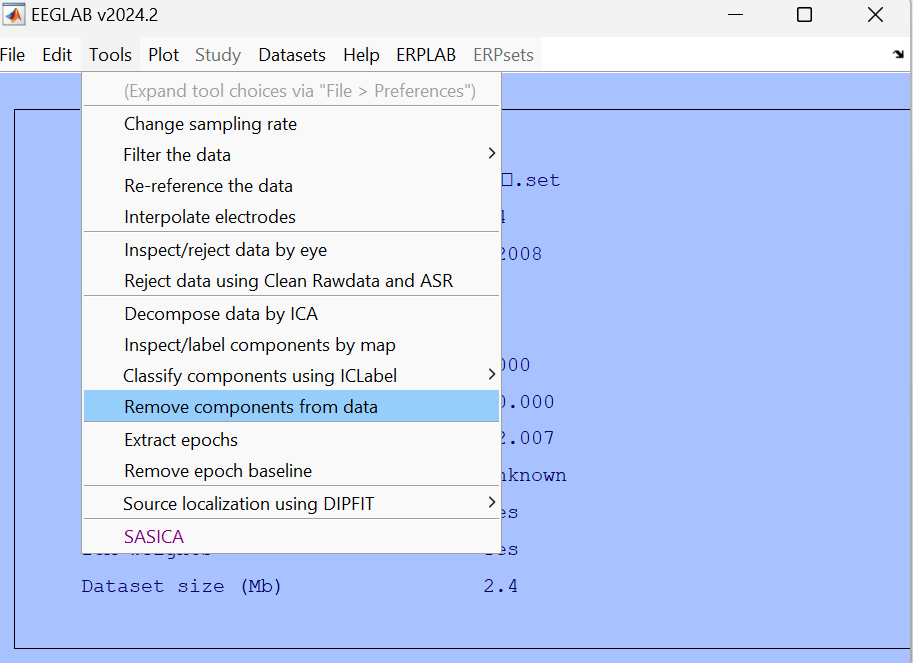
At this point, all data processing has been completed.

